Gene function prediction
The steps of the gene function prediction task are:
- Create a data set representing genes with known biological function,
- Induce a classification model from the data,
- Use the model to predict biological function of genes with unknown or partially known function.
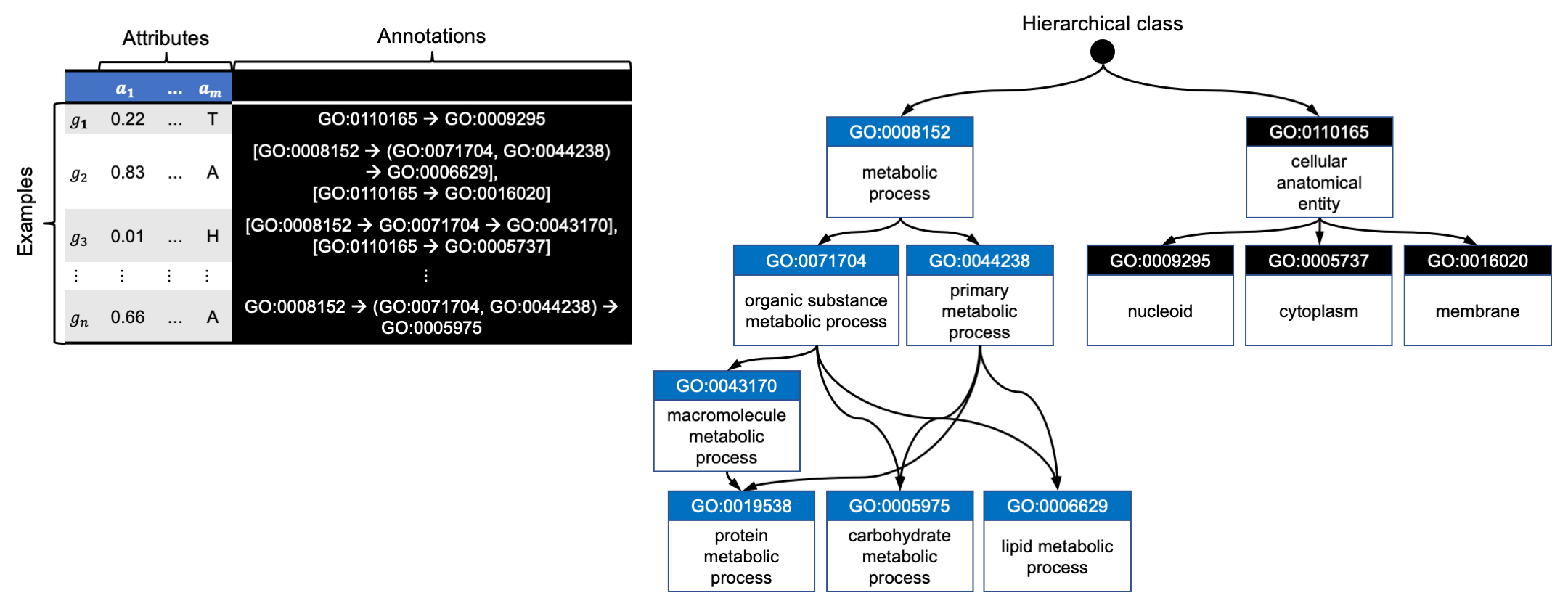
The data set has:
- Class labels that represent multiple aspects of genes' biological functions. Labels are taken from Gene Ontology and are represented with ID numbers (e.g., GO:0006629) and descriptions (e.g., lipid metabolic process).
- Labels organised in a hierarchy with nodes representing functions and relations among nodes bottom-up generalisation of gene function (e.g., membrane is a cellular anatomical entity).
- Hierarchy in the form of a directed acyclic graph, where a node can have multiple parents.
- Examples annotated with one or several paths in the hierarchy. For example:
- Gene g1 is annotated with one path, which shows that its function is manifested in a nucleoid (GO:0009295), which is a cellular anatomical entity (GO:0110165).
- Gene g2 is annotated with two paths:
- the gene participates in the lipid metabolic process: the path beginning with GO:0006629,
- the process is manifested in membrane: the path beginning with GO:0016020.